Five ways to import data into PhyloSuite
1. Search in the NCBI
Please see https://dongzhang0725.github.io/dongzhang0725.github.io/documentation/#4-3-3-1-Brief-example.
2. Download by IDs
When you are in the PhyloSuite root folder, go to example\GenBank_entries_display_and_extract folder, open 104_flatworms_mtDNA_IDs.txt file to copy the IDs.
- Open the window via
file-->Import file(s) or ID(s)in the menu bar; - Enter IDs;
- Enter your email to tell NCBI who is downloading the sequences;
- Select a work folder to deposit the sequences; selecting a work folder of
GenBank_Filecategory will download GenBank format sequence(s), selecting a work folder ofOther_Filecategory will download FASTA format sequence(s); - Click
Start.
3. Drag files to import
When you are in the PhyloSuite root folder, go to example\GenBank_entries_display_and_extract folder:
- Select a work folder in the
GenBank_Fileroot folder; - Drag
21_flatworms_mtDNA.gbfile and drop it in thedisplay area.

You can also drag a fasta file: each sequence in the file will be converted automatically to the GenBank format. Go to example\Convert_format folder and then drag cox1_NUC_mafft.fas file to the display area. The sequence name will be used to automatically fill both ID and Organism columns.
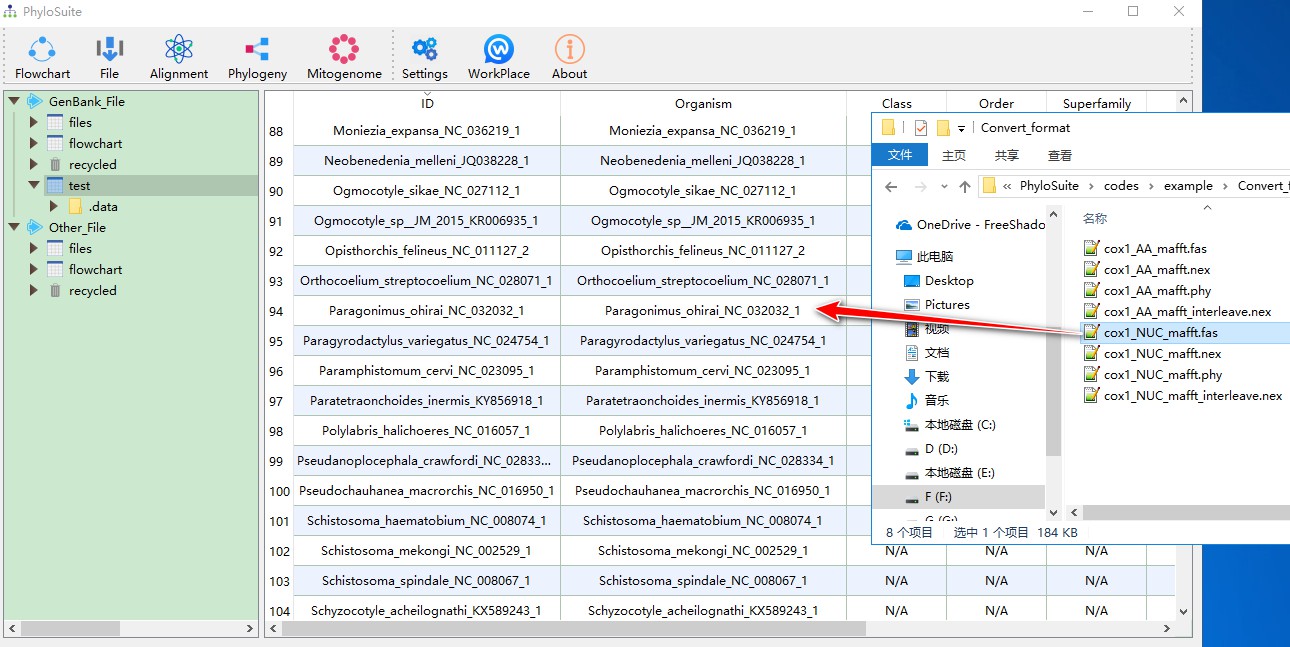
Files with sequences in fasta, phylip and nexus formats, as well as sequences recorded in a Word document, can be drag-dropped into the work folder in Other_File root folder:

4. Use button to import
Open
Inputwindow viafile-->Import file(s) or ID(s)in the menu bar, then clickChoose Filesto open files;When a work folder is empty, you can click
Open file(s)/Input ID(s)in the display area to open files.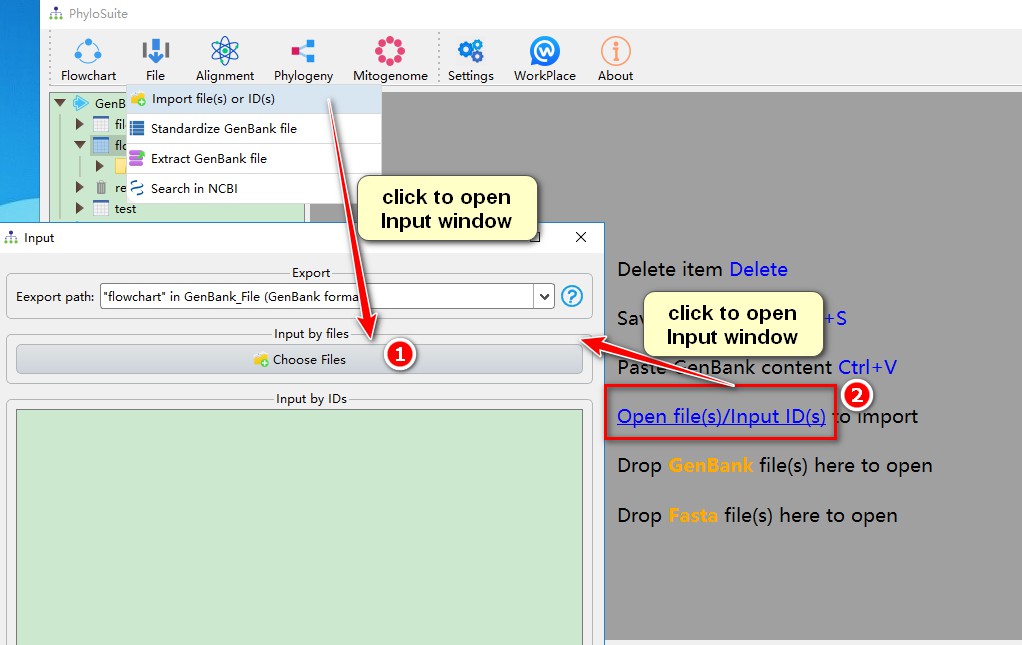
When there are sequences in the work folder, you can use
Add fileoption, accessed via the right-click menu.
5. Copy to import
Select a work folder in the
GenBank_Fileroot folder;Open
21_flatworms_mtDNA.gbfile (see 3.3),CTRL+Ato select all the contents,CTRL+Cto copy, thenCTRL+Vto paste them in the display area.
Similar to step 2, you can also open
cox1_NUC_mafft.fas(see 3.3) file to copy its contents to the display area.
When you have a raw sequence, you can also copy it into the work folder.

Reversely, you can also select and copy (
CTRL+C) sequences in thedisplay area(the content will be copied to clipboard in the GenBank format).
6. Recommended reading
Other demo tutorials can be seen here.